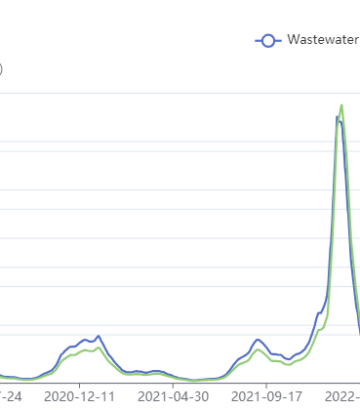
GISAID Variant Data for Canada
External link for mobile users
External link for mobile users
External link for mobile users
External link for mobile users
External link for mobile users
Data from GISAID.org1, using PANGO lineages2.
Notes on usage
- These graphs show the number of samples that are positive for a SARS-CoV-2 strain, as a percentage of the total number of samples collected on a given week whose data has been uploaded to GISAID (i.e. a variant’s prevalence).
- If a strain reaches 2% prevalence (4% on weeks with ≤50 samples), a strain’s contribution to a week’s total is shown as a segment of the week’s bar.
- If a strain does not reach 2% prevalence, its share is counted towards its parent strain (eg. EG.5.1.6’s share would be counted towards that of EG.5.1.*)
- X.X.* in the graph includes all descendants of strain X.X, except for ones that have their own segment of a given week’s bar.
- For example, if “HV.1.*” (aka. EG.5.1.6.1) and “EG.5.1.6.*” are both given segments in a week’s bar, HV.1’s prevalence would not be counted towards that of EG.5.1.6. Therefore, one must add together the percentage shares labelled “HV.1.*” and “EG.5.1.6.*” to find the weekly prevalence of all EG.5.1.6 descendant strains.
Methodology
SARS-CoV-2 sample data from each province or group of provinces is downloaded from GISAID.org. A strain’s prevalence is expressed as a percentage representing the number of samples in which the strain was detected, divided by the number of samples from the province collected in a given week. If it reaches 2% prevalence, it is given a portion of the week’s bar which may be colored the same as other strains sharing its ancestry (eg. FD.2, being an XBB.1.5 descendant, is given the same color as XBB.1.5). If the strain is gaining prominence elsewhere, as XBB.1.16 was doing in the United States during the first half of 2023, it is given its own color, which would still reflect the strain’s ancestry (eg. all recombinants are given warm colors whereas non-recombinant strains are given cold colors, as of May, 2023).
If a strain doesn’t cross the 2% threshold, it is rolled in to its parent strain, which would be the cell directly to the left of it on the phylogeny. If it and its parent strain combined do not cross the 2% threshold, the process is repeated, until their portion of the week’s samples is rolled into a category that crosses the 2% threshold. If no such category can be found, it is classed as “other recombinants”, or “other”. For example, FD.2 would be rolled into XBB.1.5.15, which would in turn be rolled into XBB.1.5.
On weeks when a single detection of a certain variant is enough to cross the 2% threshold, the threshold becomes 4% to ensure that only widely circulating variants are reported.
Due to delays in reporting sequencing data to GISAID, which are in some cases over 2 months long, only the last 4 weeks of sequences (not including the most recent week; i.e. the 2nd to 5th bars counting from the right-most one) will be reviewed for updates.
As of May, 2023, all “other” strains are omicron descendants.
References
1 Shu, Y. and McCauley, J. (2017) GISAID: from vision to reality. EuroSurveillance, 22(13). doi: 10.2807/1560-7917.ES.2017.22.13.30494. PMCID: PMC5388101
2 Roemer, C. and COV-Lineages (2023) lineage_notes.txt. https://github.com/cov-lineages/pango-designation/blob/master/lineage_notes.txt
Acknowledgments
We gratefully acknowledge all data contributors, i.e., the Authors and their Originating laboratories responsible for obtaining the specimens, and their Submitting laboratories for generating the genetic sequence and metadata and sharing via the GISAID Initiative, on which this research is based.